|
||||||||
 |
||||||||
|
|
||||||||
 |
 |
|||||||
THE TORONTO CENTRE FOR PHENOGENOMICS
MOUSE IMAGING CENTRE |
 |
|
|
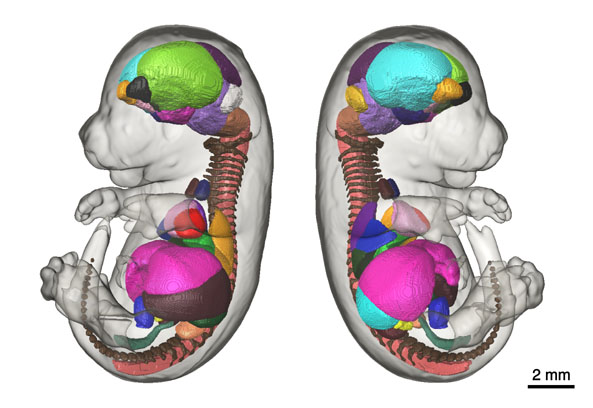
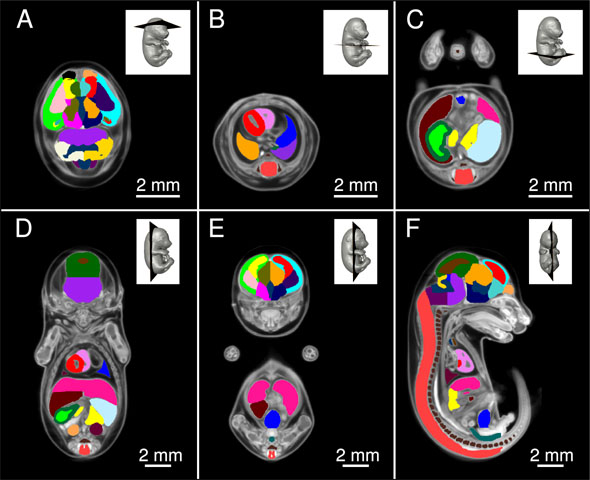
Micro-CT E15.5 Mouse Embryo AtlasThe goal of the International Mouse Phenotyping Consortium (IMPC) is to phenotype targeted knockout mouse strains throughout the whole mouse genome (23,000 genes) by 2021. A significant percentage of the generated mice will be embryonic lethal, therefore, phenotyping methods tuned to the mouse embryo are needed. There are many assays that would contribute to phenotyping these mice, however, methods that are robust, quantitative, automated and high-throughput are attractive due to the numbers of mice. 3D imaging is a useful method for characterizing phenotypes in mouse embryo morphology. However, tools to automatically quantify morphological information of mouse embryos from 3D imaging have not been fully developed. We present a representative mouse embryo average 3D atlas comprised of Micro-CT images of 35 individual C57Bl/6J mouse embryos at 15.5 days post coitum. The 35 Micro-CT images were registered into a consensus average image with our automated image registration software and 48 anatomical structures were segmented manually. A detailed description of this atlas can be found in: Wong, M. D., Dorr, A. E., Walls, J. R., Lerch, J. P., & Henkelman, R. M. (2012). A novel 3D mouse embryo atlas based on micro-CT. Development, 139, 3248–3256. doi:10.1242/dev.082016 Average of 35 Micro-CT scans used for Segmentation These images are in MINC file format and can be loaded in MNI Display software available freely from the McConnell Brain Imaging Centre at the Montreal Neurological Institute. The following webpage provides information on how to install the MINC Toolkit (Ubuntu, Debian, RPM, MacOS X); a full suite of software to manipulate and visualize MINC files: Installing the MINC Toolkit The anatomical average and labels/segmentation file in NIfTI format can be found here: files in NIfTI format
|
|
© 2004 The Centre for Phenogenomics |
|
 Back to Mouse Atlas
Back to Mouse Atlas